
1
Data Format Standardization and
DICOM Integration for Hyperpolarized
13
C MRI
Ernesto Diaz
1
, Renuka Sriram
1
, Jeremy W. Gordon
1
, Avantika Sinha
1
, Xiaoxi Liu
1
, Sule Sahin
1
,
Jason Crane
1
, Marram P Olson
1
, Hsin-Yu Chen
1
, Jenna Bernard
1
, Daniel B. Vigneron
1,2
, Zhen
Jane Wang
1
, Duan Xu
1,2
, Peder E. Z. Larson
1,2
Affiliations:
1
Department of Radiology and Biomedical Imaging, University of California – San Francisco,
San Francisco, California, USA
2
UC Berkeley-UCSF Graduate Program in Bioengineering, University of California, Berkeley and
University of California, San Francisco, California, USA
Corresponding author address:
pede[email protected], 1700 4
th
Street, Byers Hall Room 102C, San Francisco, CA 94143
Submitted to the Journal of Imaging Informatics in Medicine as a Technical Note
Keywords: Hyperpolarized
13
C MRI, metabolic imaging, DICOM format, Experiment metadata

2
Statements and Declarations
Funding
This work was supported by NIH grants P41EB013598, U24CA253377, R01CA262630, and
R01CA249909. Authors PEZL, DBV, and JWG received research support from GE Healthcare.
Competing Interests
Financial interests: Authors ED, RS, AS, XL, JB, ZJW, and XU declare they have no financial
interests. Authors PEZ, DBV, and JWG received research funding from GE Healthcare
Author Contributions
All authors contributed to the study conception and design. DICOM tools were created by
Ernesto Diaz, Jeremy Gordon, Jason Crane, Marram P Olson, and Xiaoxi Liu. Data collection
and analysis were performed by Ernesto Diaz. Creation of the proposed parameters was
performed by Peder Larson. The first draft of the manuscript was written by Ernesto Diaz and
Peder Larson, and all authors commented on previous versions of the manuscript. All authors
read and approved the final manuscript.
Ethics approval
This manuscript presents technical methods but does not include any in vivo study results.
Consent to participate
Not applicable
Consent to publish
Not applicable
3
Abstract
Hyperpolarized (HP)
13
C MRI has shown promise as a valuable modality for in vivo
measurements of metabolism and is currently in human trials at 15 research sites worldwide.
With this growth it is important to adopt standardized data storage practices as it will allow sites
to meaningfully compare data.
In this paper we (1) describe data that we believe should be stored and (2) demonstrate
pipelines and methods that utilize the Digital Imaging and Communications in Medicine
(DICOM) standard. This includes proposing a set of minimum set of information that is specific
to HP
13
C MRI studies. We then show where the majority of these can be fit into existing DICOM
Attributes, primarily via the “Contrast/Bolus” module.
We also demonstrate pipelines for utilizing DICOM for HP
13
C MRI. DICOM is the most common
standard for clinical medical image storage and provides the flexibility to accommodate the
unique aspects of HP
13
C MRI, including the HP agent information but also spectroscopic and
metabolite dimensions. The pipelines shown include creating DICOM objects for studies on
human and animal imaging systems with various pulse sequences. We also show a python-
based method to efficiently modify DICOM objects to incorporate the unique HP
13
C MRI
information that is not captured by existing pipelines. Moreover, we propose best practices for
HP
13
C MRI data storage that will support future multi-site trials, research studies and technical
developments of this imaging technique.
4
Introduction
Hyperpolarized (HP)
13
C MRI has shown promise as a valuable modality particularly for in vivo
measurements of metabolism
1
, and is currently in human trials at 15 research sites worldwide
with over 75 human research publications
2
. It is based on intravenous injection of a
hyperpolarized contrast agent that has been enriched with
13
C. The most widely used agent is
13
C-pyruvate for measuring metabolism, but there are also a range of other promising agents
undergoing clinical translation including
13
C-urea (perfusion)
3
,
13
C-alpha-ketoglutarate
(metabolism)
4
, and
13
C-fumarate (necrosis)
5
. To date, human studies have been performed to
characterize normal metabolism, and clinical studies have been performed to study in altered
metabolism in prostate cancer, brain tumors, kidney tumors, pancreatic cancer, metastatic
disease, liver disease, and heart disease
2
.
As HP
13
C MRI continues to grow, it is important to adopt standardized data storage practices,
allowing sites to compare data. The Digital Imaging and Communications in Medicine (DICOM)
format is an attractive standard format as it is commonly used in digital imaging, medical
imaging, and communications. The DICOM file type consists of the image and the metadata of
the image packed into a single file. The information in the metadata is organized as a constant
and is standardized by a series of data elements. By extracting the data elements, we can
access important information regarding the patient demographics and study parameters that are
crucial for study interpretation. The DICOM standard is actively supported and updated. It also
supports the interchange of information between computer systems such as picture archiving
and communication system (PACS).
The goal of this work is to describe an approach for HP
13
C MRI data standardization utilizing
the DICOM format. We first describe the data requirements, focusing on the unique aspects of
HP
13
C MRI. We then describe current processing pipelines that support the creation of DICOM
objects from multiple vendors and pulse sequences. Finally, we demonstrate how this
information can be incorporated into DICOM format.
Hyperpolarized
13
C MRI Data
The data requirements for HP
13
C MRI are outlined briefly below. This includes performing
imaging to spatially resolve signals, encoding of multiple metabolites to measure metabolic
conversion, performing dynamic measurements to capture the rapid kinetics, and characteristics
of the HP agent, all of which that will affect the results and analysis.
Imaging
HP
13
C experiments can be spatially resolved using MRI techniques, providing important
localization to visualize metabolic activity within the body. Therefore, a format to store multi-
dimensional data needs to be supported.
Metabolite encoding
HP
13
C MRI has the unique ability to encode signals from multiple metabolites, distinguishing
the injected agent, or substrate, from resulting metabolic products that are converted in vivo.
The most common combination of metabolites is [1-
13
C]pyruvate (substrate), [1-
13
C]lactate
(product), [1-
13
C]alanine (product), and
13
C-bicarbonate (product). Thus, the data should support
a metabolite encoding dimension. Metabolite encoding in the acquisition is typically done with
MR spectroscopy methods, chemical shift encoding (e.g. IDEAL) methods, or metabolite-

5
specific imaging
6
. The data must support metabolite encoding through identification of the
individual compounds and/or the resonant frequencies, or with a spectroscopy dimension.
Dynamic Measurements
In a HP
13
C MRI experiment, rapid metabolite kinetics following injection, including perfusion
and metabolic conversion, that are typically captured with time-resolved imaging. Thus, the data
should support dynamic measurements and record timings of the measurements.
HP
13
C Agents
To analyze HP
13
C data, it is important to know the characteristics of the HP
13
C agent, how it
was prepared, and how it was used. Table 1 lists such relevant HP
13
C agent information. Thus,
the data should support capturing of this information.
HP
13
C Agent Information
Purpose/Description
HP
13
C agent(s) injected
Describe the composition of the injected agents. Note
that this can include simultaneous injection of multiple
agents
Administration route of agent
How was the HP solution delivered
Total Injected Volume
How much HP solution was injected in total
Concentration of each HP agent
Concentration of each HP agent in HP solution
Start Time of injection
Describe the timing of the start of injection, important to
know relative to the data acquisition timing
Total injection duration
Over how long was the HP solution injected
Polarization
What was the measured polarization of the HP agent(s).
Typically back-calculated to the time of dissolution.
Polarization Measurement Timing
timing of the polarization measurement relative to the data
acquisition
Agent Relaxation Rates (e.g. T1)
Expected or measured relaxation rates used to calculate
polarization
Dissolution Timing
Timing of the dissolution of the HP agent, most important
to know relative to the data acquisition
Table 1:Hyperpolarized
13
C agent information that we typically keep track of during our studies.
DICOM Processing Pipelines
Our current processing pipelines support DICOM creation from multiple vendors and pulse
sequences, listed in Table 2. The spectroscopy sequences use an open-source standards-
based software framework(SIVIC) for DICOM creation which includes support for DICOM
Spectroscopy
(7,8).

6
Table 2: Currently supported sequences with DICOM creation pipelines in use at our institution
in alphabetical order.
MR Spectroscopy and Spectroscopic Imaging with SIVIC
MR spectroscopy and spectroscopic imaging (MRS/I) for HP
13
C MRI includes data sampling at
various time delays to resolve a spectrum and separate
13
C-labeled compounds. For processing
of MRS/I data, we utilize the libraries in SIVIC for data processing and DICOM creation,
including support of the DICOM MRS standard
9
. This software can reads in multiple vendor
MRS raw data files (Bruker, GE, Philips, Siemens, Varian), and it for HP
13
C MRI studies on GE
Healthcare and Bruker MRI scanners.
1. Reads in raw data
2. Performs MR spectroscopy or MR spectroscopic imaging reconstructions
Scanner Vendor
Sequences
Raw Data
Tools
Output
Bruker
Slab-selective
MRS, chemical
shift imaging
(CSI) and Echo-
planar
spectroscopic
imaging
MRS Raw
File or Dad
File
SIVIC
MATLAB
MR
Spectroscopy
DICOMs
Bruker
Metabolite-
Specific
Imaging
FID
Bruker Paravision
MATLAB
Metabolite
image, AUC,
kinetic rate
DICOMs
GE Healthcare
Slab-selective
MRS, chemical
shift imaging
(CSI) and Echo-
planar
spectroscopic
imaging
Pfile or
ScanArchive
SIVIC
MR
Spectroscopy
DICOMs
GE Healthcare
Metabolite-
specific EPI
Pfile or
ScanArchive
GE Orchestra
Toolbox
MATLAB
Metabolite
images, AUC
ratio and kinetic
rate DICOMs
RTHawk Research
(Vista.ai)
Metabolite-
specific spiral
and bSSFP
RTHawk raw
data
Real-time
reconstructed
DICOM
MATLAB to improve
data
Metabolite
image DICOMs

7
3. Writes to DICOM MRS file
4. Optional – extract peak amplitudes of areas from spectra and create individual
metabolite DICOM files
5. Optional - write raw data or partially processed data to DICOM
6. Optional - secondary capture of MRS and anatomical visualization to DICOM
GE Metabolite-specific Imaging
Metabolite-specific imaging for HP
13
C MRI uses a spectral-spatial RF pulse to selectively excite
a single metabolite
10
and is followed by fast imaging readout such as echo-planar imaging (EPI)
or spirals. We have implemented EPI-based metabolite-specific imaging for GE Healthcare MRI
scanners along with a reconstruction pipeline based on the GE Orchestra reconstruction
toolbox
11
. This leverages the EPI processing and DICOM creation functionality that is utilized by
GE product pulse sequences.
1. Reads in raw data (Pfile or ScanArchive)
2. Perform EPI reconstruction including Nyquist ghost correction
3. Perform HP-specific coil combination
12
4. Create area-under-curve (AUC) and kinetic rate maps
5. Write metabolite images and parameter maps to DICOM
Figure 1: Example of how HP
13
C MRI can be used in a workflow for improved targeting of
biopsies in prostate cancer
11
. Using DICOM has been critical in the success of this workflow, as
this allows for integration into PACS as well as the fusion biopsy platform, with radiologists and
urologists easily able to review the images and resulting annotations outlining the biopsy
targets. Figure reproduced with permission from
11
.
RTHawk Metabolite-specific Imaging
We have implemented metabolite-specific imaging with EPI and spirals, as well as metabolite-
specific balanced steady-state free-precession (bSSFP) sequences
13-15
on the RTHawk
Research platform. This is a vendor-neutral platform that allows for advanced control of the

8
MRI system including real-time reconstructions and feedback as well as pulse sequence
programming.
1. We utilize real-time reconstruction built into RTHawk during the scanning process,
resulting in individual metabolite DICOM images at each timepoint.(Note: This uses the
sum of squares coil combination technique.)
2. The DICOMs Images are exported when the data is transferred from the RTHawk
workstation to the local network.
3. A second, improved image reconstruction is performed using raw complex-valued data
and a more advanced coil combination method to enhance the data quality
9
.
4. To integrate the improved data into DICOM, a MATLAB script is employed that reads the
metadata from the real-time reconstructed DICOMs Images, replaces the original voxel
data with the results of the second, optimized reconstruction, and creates new DICOMs
Images with appropriate new unique identifiers (UIDs).
Bruker Metabolite-specific Imaging
Similar to the GE metabolite-specific imaging sequence, it uses a spectral-spatial RF pulse to
excite a single metabolite
10
, and is followed by a fast echo-planar imaging (EPI) readout
16
.
1. Bruker scanner has its own built-in EPI reconstruction and an option to write
reconstructed metabolite maps to DICOM files.
2. A MATLAB script is used to read in the DICOM files. From that data, we generate AUC
maps and kinetic rate maps, and write back to DICOM files.
3. Optional: Reads in Bruker raw data using a MATLAB script, use a custom reconstruction
script and write to DICOM.
DICOM HP Agent Integration
Many of the HP agent metadata that are desirable for analyses (Table 1) can be integrated into
existing DICOM attributes, particularly within the “Contrast/Bolus” module. However, the above
DICOM pipelines by default do not include support for these metadata. To address this, we built
a tool to modify HP
13
C DICOM objects based on user input to include this HP agent metadata.
Currently, the HP
13
C images created by MRI scanners or in-house reconstruction pipelines do
not utilize the Contrast/Bolus Attributes. We seek to leverage the existing Contrast/Bolus
Attributes in DICOM to store information to identify the study and experimental parameters that
are valuable for HP
13
C data analysis by researchers and clinicians. Table 3 lists the attributes
we are adding to our images.
TAG
VR
Attribute Name
1
H MRI
Example
Values
HP
13
C Description
HP
13
C Example Values
(0018,0010)
LO
Contrast/Bolus Agent
CC
MAGNEVIST
ORAL &
OMNIPAQUE
HP
13
C agent(s)
injected
HYPERPOLARIZED [1-
13C]PYRUVATE
HYPERPOLARIZED [2-
13C]PYRUVATE

9
HYPERPOLARIZED [1-
13C]PYRUVATE +
[13C,15N]-UREA
(0018,1040)
LO
Contrast/Bolus Route
Administration route
of contrast agent
IV
(0018,1041)
DS
Contrast/Bolus
Volume
Injected volume
(mL)
(0018,1042)
TM
Contrast/Bolus Start
Time
Define start of
injection relative to
data acquisition (Not
DICOM compliant)
Start Time in
HHMMSS.FFFFFF
(DICOM compliant)
(0018,1047)
DS
Contrast/Bolus Flow
Duration
Total injection
duration [s]
(0018,1048)
CS
Contrast/Bolus
Ingredient
IODINE
BARIUM
GADOLINIUM
CARBON
DIOXIDE
13
C enriched
compounds
[1-^13^C]PYRUVATE
[2-^13^C]PYRUVATE
[^13^C,^15^N]UREA
[1-^13^C]ALPHA-
KETOGLUTARATE
(0018,1049)
DS
Contrast/Bolus
Ingredient
Concentration
Concentration of
each
13
C compound
[mg/mL]
(0400,0550)
SQ
Modified Attributes
Sequence
Store prior versions
of an attributes that
were removed or
modified
(0400,0562)
DT
Attribute Modification
DateTime
Tracked Date and
Time when
attributes were
modified
(0400,0563)
LO
Modifying System
Describes what
modified the
attributes
SIVIC-
HP_agent_DICOM_tool.py
Table 3: DICOM attributes proposed for addition to HP
13
C MRI study data. This captures the
majority but not all the desired parameters listed in Table 1.Note that the “Contrast/Bolus
Ingredient” attributes listed can include more than one entry to support multiple simultaneously
polarized HP
13
C agents
17
such as pyruvate/urea
3,14
. The proposed
13
C naming conventions are
in a style that is consistent with the style used for PET in DICOM.
In DICOM, each attribute or data element is identified by a unique tag (TAG), which is
comprised of a group number and an element number. The value representation (VR) is a code
that indicates the data type used to encode the value(s) of the attribute. Additionally, the value
multiplicity (VM) indicates how many values can be present in the attribute. These three

10
components, work together to provide a comprehensive understanding of the attribute and its
role within the DICOM file, allowing for accurate and efficient manipulation and interpretation of
the medical imaging data.
We reviewed the DICOM standards definition and found the listed attributes in Table 3 can
capture much of the unique HP agent metadata. This includes ingredients, injection timing, and
dosing characteristics. These can all be added in a DICOM compliant fashion.
One important feature captured in this proposal is the inclusion of multiple ingredients, which
can capture dual-agent (“co-polarized”) or multi-agent HP studies
3,14,17
. In these studies, multiple
13
C-enriched compounds are simultaneously hyperpolarized and simultaneously injected. The
“Contrast/Bolus Agent Sequence”, which can capture use of multiple agents that affect the data
in a DICOM object, was not used since when multiple HP compounds are desired, they are
combined into a single agent with multiple ingredients. The use of multiple agents is not
performed to our knowledge, as it would require rapid dissolution of multiple separate samples
following hyperpolarized to inject both before the signal decays.
We also proposed standardized ingredient names that are consistent with the style used in PET.
In these, the enriched isotope(s) are denoted with “^” around the nucleus, e.g. “^13^C”. We also
use a standard chemical naming convention to identify the site of enrichment within the
molecule.
We used the PyDicom python toolbox to access the data in DICOM files (Figure 2). PyDicom is
a general-purpose DICOM framework whose purpose is to reads and write DICOM attributes. It
can also add, delete, and modify any attributes to DICOM objects.
Figure 2: This is an example of the PyDicom functions used. It reads a file, then adds an
attribute and saves the file.
The basic workflow of the python script created using the packages PyDicom and OS is as
follows: User would run the script in the terminal with a parameter of a directory path for
DICOMS that need to be modified. The prompts would pop up one by one and the user can
write its own inputs or press enter for default values as shown in Figure 3. Once the user has
entered the prompts, it will loop through the folder and add all the attributes to each DICOM file.
We have also aimed to capture the modifications made to the DICOM file when the tool is used.
For this, we used the Modified Attributes Sequence to describe what attributes we modified. For
Modifying System, it shows the name of this tool we used to change the attributes. We used
DateTime within the Modified Attributes Sequence to keep track of the date when we modified
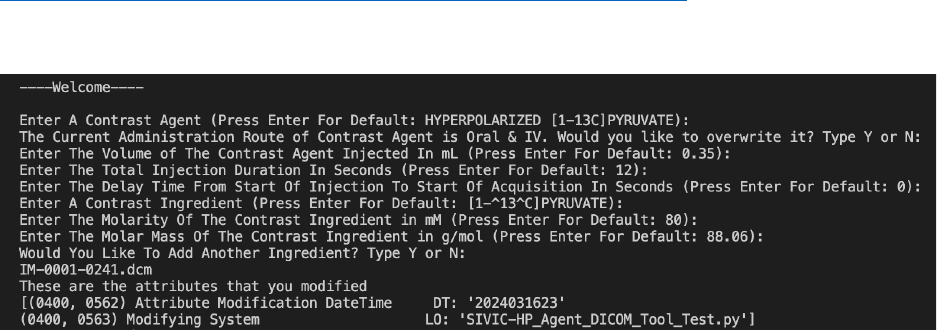
11
the attributes, and this shows other researchers when we modified them. This maybe a non-
standard use of these attributes.
This tool has been demonstrated on DICOM objects from a metabolite-specific EPI sequence
18
on a GE 3T scanner, reconstructed with a custom implementation based on the GE Orchestra
package, DICOM from Bruker 2D CSI scan generated by SIVIC, and a DICOM from a GE echo-
planar spectroscopic imaging (EPSI) sequence generated by SIVIC.
The script is available within SIVIC at
https://github.com/SIVICLab/sivic/tree/master/applications/dicom_tools.
Figure 4: The prompts questions that gives option for default values or user input. As well,
showing the attributes that has been modified.
HP
13
C MRI Data Storage Best Practices
We have also created a proposed set of Data Storage Best Practices. These were based upon
a survey of research groups at our institution, performing both human and preclinical HP
13
C
imaging studies at our institution. The proposed set of Data Storage Best Practices aim to
ensure that the stored data completely describes the study as is necessary for subsequent
processing. This should also be done so that future researchers can easily retrieve and analyze
the data without the need to search multiple sources of information.
Storage of essential study data should be done in a centralized location with high-quality back
up, using a single folder per study, including:
• HP spectroscopy/image data (DICOM recommended)
• HP raw data
•
1
H image data (DICOM recommended)
• Copy of any processing scripts
• Metadata – any other information about the study. This can either be directly noted in the
data folder, or in a separate database/spreadsheet. At a minimum this should include
o Image acquisition data (ideally in DICOM files):
o Flip angle information, especially for variable flip angle or metabolite-specific flip
schedules
o Injection timing characteristics – length of injection, timing of imaging relative to
injection(See Table 1)
o Dose characteristics – volume injected, concentration of agent, subject weight,
polarization.(See Table 1)
12
• Other Study data - directly recorded in the study folder, or in a separate
database/spreadsheet.
• Study “Key” needed in all data locations to link to other metadata sources.
o Option 1: Unique ID for each study
o Option 2: Unique ID for each subject + Unique ID for each study
Discussion and Unmet Needs
Using the DICOM standard has significant advantages for HP
13
C MRI data. Given this
technology is being translated into clinical studies, DICOM will allow for seamless integration
into existing image viewing and analysis platforms, ultimately supporting use in clinical
workflows. The use of standardized DICOM objects would also be valuable in the multi-site
clinical trials setting, which are likely to begin within the next several years.
We also believe that the DICOM format will be valuable for the preclinical studies given the
flexibility of the format. A pre-clinical DICOM standard has recently been proposed
19
, and a
recent assessment preclinical imaging metadata need has also advocated for integration into
DICOM
20
. While adoption is less consistent in preclinical imaging systems, we have
demonstrated a pipeline for our Bruker preclinical MRI scanner.
While the Contrast/Bolus module in DICOM supported most HP agent parameters we wanted to
save, there were several parameters without an existing attribute. These include the polarization
measurements, T1 relaxation rate, and dissolution timing. These could be stored in private data
elements. This could also justify the need for a HP Agent MRI Module in DICOM. This could
largely follow the examples from PET, which also must capture similar information: polarization
is analogous to specific activity, relaxation rates are analogous to half-life, and dissolution timing
is analogous to dose creation timing. The PET Isotope Module (C.8.9.2) could serve as a good
model.
Several other current features of the DICOM standard may also be relevant for HP MRI. The
Enhanced MR Information Object Definitions (IODs) are attractive because they allow for more
MRI specific data sources such as raw k-space and spectroscopy data and can be used for
“raw” images (e.g. dynamic metabolite images) as well as derived parameter maps (e.g. kinetic
rate maps). The Enhanced Contrast/Bolus Module may also be relevant to store additional HP
agent parameters.
Hyperpolarization is also used in HP
129
Xe MRI, a modality that unique images pulmonary
ventilation, gas exchange, and terminal airway morphology
22
and received FDA approval in
2021. There are many shared aspects of both HP MRI methods, including the hyperpolarization
of an agent, administration of this agent (inhaled for HP
129
Xe), relatively rapid imaging of the
agent, and measuring agent kinetics. With this in mind, we believe it would be beneficial to
coordinate efforts across the HP modalities when developing DICOM integration methods and
specifications of metadata.
HP
13
C MRS and MRSI acquisition methods are more challenging to use in a standardized
fashion compared to imaging-based methods (e.g. metabolite-specific imaging). This is because
the adoption of DICOM MRS by vendors is rather limited, leading to the community
development of tools such as SIVIC
8
. If MRS and MRSI methods continue to show value, it
13
would be important for researchers to work closely with vendor partners and ideally have the
DICOM MRS standard integrated into the scanners.
Ultimately, we believe it is important that both a standardized and essential set of DICOM
attributes are recorded for HP
13
C MRI studies. This should be standardized so as not to
introduce confusion, for example due to different naming conventions or use of different DICOM
attributes. This should also include an essential set of attributes to allow for robust and
reproducible analyses of HP
13
C MRI data. The final definitions are beyond the scope of this
paper, but we think would be best addressed by a consensus of HP
13
C MRI researchers in
future discussions as the community recently formed the HP 13C Consensus Group
2
. Following
consensus, any changes to the DICOM standard should then be proposed by drafting a
Correction Proposal (CP) and work with the DICOM MR Working Group (WG 16) to submit it to
the base standards WG (WG 6).
Conclusion
We have proposed a set of attributes to store HP
13
C MRI metadata, particularly regarding the
HP agent, and implemented via PyDicom a program to add these a set of these attributes in a
DICOM-compliant fashion based on user input. This was shown to work in line with a variety of
DICOM creation pipelines that are used for HP
13
C data. In addition, we have proposed a
broader set of data storage best practices. Providing further data standardization will advance
this technology by allowing sites to seamlessly share and compare data, including in the context
of multi-site trials, and by using DICOM HP
13
C MRI can be easily integrated into other clinical
and research workflows.
14
References
1. Wang ZJ, Ohliger MA, Larson PEZ, et al. Hyperpolarized 13C MRI: State of the Art and
Future Directions. Radiology. Published online March 5, 2019:182391.
doi:10.1148/radiol.2019182391
2. Larson PE, Bernard JM, Bankson JA, et al. Current Methods for Hyperpolarized [1-
13C]pyruvate MRI Human Studies. Published online September 7, 2023.
doi:10.48550/arXiv.2309.04040
3. Qin H, Tang S, Riselli AM, et al. Clinical translation of hyperpolarized 13C pyruvate and urea
MRI for simultaneous metabolic and perfusion imaging. Magnetic Resonance in Medicine.
n/a(n/a). doi:10.1002/mrm.28965
4. Chaumeil MM, Larson PEZ, Yoshihara HAI, et al. Non-invasive in vivo assessment of IDH1
mutational status in glioma. Nat Commun. 2013;4:2429. doi:10.1038/ncomms3429
5. Clatworthy MR, Kettunen MI, Hu DE, et al. Magnetic resonance imaging with hyperpolarized
[1,4-(13)C2]fumarate allows detection of early renal acute tubular necrosis. Proc Natl Acad Sci
U S A. 2012;109(33):13374-13379. doi:10.1073/pnas.1205539109
6. Gordon JW, Chen H, Dwork N, Tang S, Larson PEZ. Fast Imaging for Hyperpolarized MR
Metabolic Imaging. Magnetic Resonance Imaging. 2021;53(3):686-702. doi:10.1002/jmri.27070
7. Crane JC, Olson MP, Nelson SJ. SIVIC: Open-Source, Standards-Based Software for
DICOM MR Spectroscopy Workflows. International Journal of Biomedical Imaging.
2013;2013:e169526. doi:10.1155/2013/169526
8. SIVIC. doi:10.5281/zenodo.4777197
9. Crane JC, Gordon JW, Chen HY, et al. Hyperpolarized 13C MRI data acquisition and
analysis in prostate and brain at University of California, San Francisco. NMR in Biomedicine.
2020;n/a(n/a):e4280. doi:10.1002/nbm.4280
10. Cunningham CH, Chen AP, Lustig M, et al. Pulse sequence for dynamic volumetric imaging
of hyperpolarized metabolic products. J Magn Reson. 2008;193(1):139-146.
11. Chen HY, Bok RA, Cooperberg MR, et al. Improving multiparametric MR-transrectal
ultrasound guided fusion prostate biopsies with hyperpolarized 13 C pyruvate metabolic
imaging: A technical development study. Magn Reson Med. 2022;88(6):2609-2620.
doi:10.1002/mrm.29399
12. Zhu Z, Zhu X, Ohliger MA, et al. Coil combination methods for multi-channel hyperpolarized
13C imaging data from human studies. Journal of Magnetic Resonance. 2019;301:73-79.
doi:10.1016/j.jmr.2019.01.015
13. Tang S, Bok R, Qin H, et al. A metabolite-specific 3D stack-of-spiral bSSFP sequence for
improved lactate imaging in hyperpolarized [1-13C]pyruvate studies on a 3T clinical scanner.
Magnetic Resonance in Medicine. n/a(n/a). doi:10.1002/mrm.28204
14. Liu X, Tang S, Mu C, et al. Development of specialized magnetic resonance acquisition
techniques for human hyperpolarized [13 C,15 N2 ]urea + [1-13 C]pyruvate simultaneous
15
perfusion and metabolic imaging. Magn Reson Med. 2022;88(3):1039-1054.
doi:10.1002/mrm.29266
15. Liu X, Tang S, Cui D, et al. A metabolite specific 3D stack-of-spirals bSSFP sequence for
improved bicarbonate imaging in hyperpolarized [1-13C]Pyruvate MRI. J Magn Reson.
2023;353:107518. doi:10.1016/j.jmr.2023.107518
16. Sahin SI, Ji X, Agarwal S, et al. Metabolite-Specific Echo Planar Imaging for Preclinical
Studies with Hyperpolarized 13C-Pyruvate MRI. Tomography. 2023;9(2):736-749.
doi:10.3390/tomography9020059
17.Wilson DM, Keshari KR, Larson PEZ, et al. Multi-compound Polarization by DNP Allows
Simultaneous Assessment of Multiple Enzymatic Activities In Vivo. J Magn Reson.
2010;205(1):141-147.
18.Gordon JW, Vigneron DB, Larson PEZ. Development of a symmetric echo planar imaging
framework for clinical translation of rapid dynamic hyperpolarized (13) C imaging. Magn Reson
Med. Published online February 2016. doi:10.1002/mrm.26123
19. Kalen JD, Clunie DA, Liu Y, Tatum JL, Jacobs PM, Kirby J, Freymann JB, Wagner U,
Smith KE, Suloway C, et al. Design and Implementation of the Pre-Clinical DICOM Standard in
Multi-Cohort Murine Studies. Tomography. 2021; 7(1):1-9.
https://doi.org/10.3390/tomography7010001
20. Moore SM, Quirk JD, Lassiter AW, Laforest R, Ayers GD, Badea CT, Fedorov AY, Kinahan
PE, Holbrook M, Larson PEZ, et al. Co-Clinical Imaging Metadata Information (CIMI) for Cancer
Research to Promote Open Science, Standardization, and Reproducibility in Preclinical
Imaging. Tomography. 2023; 9(3):995-1009. https://doi.org/10.3390/tomography9030081
21. Niedbalski PJ, Hall CS, Castro M, et al. Protocols for multi-site trials using hyperpolarized
129Xe MRI for imaging of ventilation, alveolar-airspace size, and gas exchange: A position
paper from the 129Xe MRI clinical trials consortium. Magn Reson Med. 2021; 86: 2966–2986.
https://doi.org/10.1002/mrm.28985
